
 |  |
| Residue number and Amino Acid | Probabilities | ||
| Inside | Membrane | Outside | |
| 1 M | 0.32263 | 0.00000 | 0.67738 |
| 2 G | 0.32263 | 0.00000 | 0.67738 |
| 3 Y | 0.32263 | 0.00000 | 0.67738 |
| 4 P | 0.32263 | 0.00000 | 0.67738 |
| 5 D | 0.32263 | 0.00000 | 0.67738 |
| 6 N | 0.32263 | 0.00000 | 0.67738 |
| 7 A | 0.32263 | 0.00000 | 0.67738 |
| 8 M | 0.32263 | 0.00000 | 0.67738 |
| 9 A | 0.32263 | 0.00000 | 0.67738 |
| 10 A | 0.32263 | 0.00000 | 0.67738 |
| 11 G | 0.32263 | 0.00000 | 0.67738 |
| 12 E | 0.32263 | 0.00000 | 0.67738 |
| 13 Q | 0.32263 | 0.00000 | 0.67738 |
| 14 V | 0.32263 | 0.00000 | 0.67738 |
| 15 V | 0.32263 | 0.00000 | 0.67737 |
| 16 L | 0.32263 | 0.00000 | 0.67737 |
| 17 H | 0.32263 | 0.00000 | 0.67737 |
| 18 R | 0.32263 | 0.00000 | 0.67737 |
| 19 H | 0.32258 | 0.00006 | 0.67736 |
| 20 P | 0.31945 | 0.00885 | 0.67169 |
| 21 H | 0.30250 | 0.02586 | 0.67165 |
| 22 W | 0.30017 | 0.03175 | 0.66808 |
| 23 T | 0.29546 | 0.08489 | 0.61965 |
| 24 R | 0.21921 | 0.34695 | 0.43383 |
| 25 L | 0.07687 | 0.73651 | 0.18662 |
| 26 V | 0.03390 | 0.91178 | 0.05432 |
| 27 W | 0.01161 | 0.97002 | 0.01837 |
| 28 P | 0.00785 | 0.97845 | 0.0137 |
| 29 V | 0.00076 | 0.99758 | 0.00167 |
| 30 V | 0.00021 | 0.99943 | 0.00036 |
| 31 V | 0.00006 | 0.99982 | 0.00012 |
| 39 A | 0.00005 | 0.99975 | 0.0002 |
| 40 A | 0.00010 | 0.99946 | 0.00044 |
| 41 L | 0.00017 | 0.99880 | 0.00103 |
| 42 G | 0.00125 | 0.99371 | 0.00504 |
| 43 S | 0.00956 | 0.97743 | 0.01301 |
| 44 G | 0.01137 | 0.95605 | 0.03258 |
| 45 F | 0.01946 | 0.94365 | 0.03689 |
| 46 V | 0.12089 | 0.83446 | 0.04465 |
| 47 N | 0.39021 | 0.40204 | 0.20776 |
| 48 S | 0.56705 | 0.14464 | 0.28831 |
| 49 T | 0.65631 | 0.02931 | 0.31438 |
| 50 Q | 0.67289 | 0.00974 | 0.31737 |
| 51 W | 0.67349 | 0.00864 | 0.31787 |
| 52 Q | 0.67732 | 0.00040 | 0.32228 |
| 53 Q | 0.67692 | 0.00053 | 0.32255 |
| 54 L | 0.66801 | 0.00940 | 0.3226 |
| 55 A | 0.66353 | 0.01388 | 0.3226 |
| 56 K | 0.66243 | 0.01503 | 0.32254 |
| 57 N | 0.63478 | 0.04603 | 0.31919 |
| 58 I | 0.33721 | 0.36291 | 0.29989 |
| 59 I | 0.24236 | 0.46060 | 0.29704 |
| 60 H | 0.14735 | 0.56075 | 0.2919 |
| 61 G | 0.07413 | 0.79762 | 0.12825 |
| 62 V | 0.06090 | 0.88375 | 0.05535 |
| 63 I | 0.03636 | 0.93546 | 0.02817 |
| 64 W | 0.02923 | 0.94680 | 0.02398 |
| 65 G | 0.02622 | 0.95213 | 0.02166 |
| 66 I | 0.01615 | 0.97409 | 0.00975 |
| 67 W | 0.00352 | 0.99501 | 0.00147 |
| 68 L | 0.00032 | 0.99890 | 0.00078 |
| 69 V | 0.00008 | 0.99963 | 0.00029 |
| 72 G | 0.00005 | 0.99975 | 0.0002 |
| 73 W | 0.00005 | 0.99973 | 0.00023 |
| 74 L | 0.00005 | 0.99930 | 0.00065 |
| 75 T | 0.00008 | 0.99838 | 0.00155 |
| 76 L | 0.00012 | 0.99458 | 0.0053 |
| 77 W | 0.00033 | 0.97870 | 0.02096 |
| 78 P | 0.00134 | 0.85938 | 0.13928 |
| 79 F | 0.00144 | 0.84730 | 0.15126 |
| 80 L | 0.00263 | 0.80404 | 0.19333 |
| 81 S | 0.01610 | 0.58144 | 0.40246 |
| 82 W | 0.01912 | 0.51322 | 0.46766 |
| 83 L | 0.02552 | 0.44188 | 0.5326 |
| 84 T | 0.20328 | 0.20324 | 0.59348 |
| 85 T | 0.26086 | 0.13264 | 0.6065 |
| 86 H | 0.28185 | 0.08558 | 0.63257 |
| 87 F | 0.28404 | 0.07602 | 0.63994 |
| 88 V | 0.28734 | 0.06941 | 0.64325 |
| 89 V | 0.30023 | 0.04640 | 0.65337 |
| 90 T | 0.31693 | 0.01276 | 0.67032 |
| 91 N | 0.32183 | 0.00161 | 0.67656 |
| 92 R | 0.32262 | 0.00041 | 0.67697 |
| 93 R | 0.32268 | 0.00035 | 0.67697 |
| 94 V | 0.32268 | 0.00301 | 0.6743 |
| 95 M | 0.32272 | 0.00344 | 0.67384 |
| 96 Y | 0.32277 | 0.00355 | 0.67368 |
| 97 R | 0.32293 | 0.00347 | 0.6736 |
| 98 Q | 0.32294 | 0.00356 | 0.6735 |
| 99 G | 0.32284 | 0.00549 | 0.67167 |
| 100 V | 0.32281 | 0.00613 | 0.67106 |
| 101 L | 0.32280 | 0.00622 | 0.67098 |
| 102 T | 0.32280 | 0.00622 | 0.67097 |
| 103 R | 0.32280 | 0.00623 | 0.67097 |
| 104 S | 0.32280 | 0.00623 | 0.67096 |
| 105 G | 0.32280 | 0.00623 | 0.67096 |
| 106 I | 0.32280 | 0.00623 | 0.67096 |
| 107 D | 0.32280 | 0.00623 | 0.67096 |
| 108 I | 0.32280 | 0.00623 | 0.67096 |
| 109 P | 0.32280 | 0.00623 | 0.67096 |
| 110 L | 0.32280 | 0.00623 | 0.67096 |
| 111 A | 0.32281 | 0.00623 | 0.67096 |
| 112 R | 0.32287 | 0.00616 | 0.67097 |
| 113 I | 0.32289 | 0.00614 | 0.67097 |
| 114 N | 0.32304 | 0.00598 | 0.67099 |
| 115 S | 0.32312 | 0.00589 | 0.67099 |
| 116 V | 0.32324 | 0.00577 | 0.67099 |
| 117 E | 0.32798 | 0.00092 | 0.6711 |
| 118 F | 0.32806 | 0.00084 | 0.6711 |
| 119 R | 0.32879 | 0.00008 | 0.67113 |
| 120 D | 0.32885 | 0.00001 | 0.67113 |
| 121 R | 0.32885 | 0.00001 | 0.67113 |
| 122 I | 0.32885 | 0.00001 | 0.67113 |
| 123 F | 0.32885 | 0.00001 | 0.67114 |
| 124 E | 0.32886 | 0.00000 | 0.67114 |
| 125 R | 0.32886 | 0.00001 | 0.67114 |
| 126 M | 0.32885 | 0.00001 | 0.67114 |
| 127 L | 0.32885 | 0.00001 | 0.67114 |
| 128 R | 0.32885 | 0.00001 | 0.67114 |
| 129 T | 0.32885 | 0.00001 | 0.67114 |
| 130 G | 0.32885 | 0.00001 | 0.67114 |
| 131 T | 0.32885 | 0.00001 | 0.67114 |
| 132 L | 0.32885 | 0.00001 | 0.67114 |
| 133 I | 0.32885 | 0.00001 | 0.67114 |
| 134 I | 0.32885 | 0.00001 | 0.67114 |
| 135 E | 0.32885 | 0.00001 | 0.67114 |
| 136 S | 0.32885 | 0.00001 | 0.67114 |
| 137 A | 0.32885 | 0.00001 | 0.67114 |
| 138 S | 0.32885 | 0.00001 | 0.67114 |
| 139 Q | 0.32885 | 0.00001 | 0.67114 |
| 140 D | 0.32885 | 0.00001 | 0.67114 |
| 141 P | 0.32885 | 0.00001 | 0.67114 |
| 142 L | 0.32885 | 0.00001 | 0.67114 |
| 143 E | 0.32885 | 0.00001 | 0.67114 |
| 144 F | 0.32885 | 0.00001 | 0.67114 |
| 145 Y | 0.32885 | 0.00001 | 0.67114 |
| 146 N | 0.32885 | 0.00001 | 0.67114 |
| 147 I | 0.32885 | 0.00001 | 0.67114 |
| 148 P | 0.32886 | 0.00000 | 0.67114 |
| 149 R | 0.32886 | 0.00000 | 0.67114 |
| 150 L | 0.32886 | 0.00000 | 0.67114 |
| 151 R | 0.32886 | 0.00000 | 0.67114 |
| 152 E | 0.32886 | 0.00000 | 0.67114 |
| 153 V | 0.32886 | 0.00000 | 0.67114 |
| 154 H | 0.32886 | 0.00000 | 0.67114 |
| 155 A | 0.32886 | 0.00000 | 0.67114 |
| 156 L | 0.32886 | 0.00000 | 0.67114 |
| 157 L | 0.32886 | 0.00000 | 0.67114 |
| 158 Y | 0.32886 | 0.00000 | 0.67114 |
| 159 H | 0.32886 | 0.00000 | 0.67114 |
| 160 E | 0.32886 | 0.00000 | 0.67114 |
| 161 V | 0.32886 | 0.00000 | 0.67114 |
| 162 F | 0.32886 | 0.00000 | 0.67114 |
| 163 D | 0.32886 | 0.00000 | 0.67114 |
| 164 T | 0.32886 | 0.00000 | 0.67114 |
| 165 L | 0.32886 | 0.00000 | 0.67114 |
| 166 G | 0.32886 | 0.00000 | 0.67114 |
| 167 S | 0.32886 | 0.00000 | 0.67114 |
| 168 E | 0.32886 | 0.00000 | 0.67114 |
| 169 E | 0.32886 | 0.00000 | 0.67114 |
| 170 A | 0.32886 | 0.00000 | 0.67114 |
| 171 P | 0.32886 | 0.00000 | 0.67114 |
| 172 S | 0.32886 | 0.00000 | 0.67114 |
| General Statistics |
| TMHMM predicts transmembrane helices and the location of the intervening loop regions. |
| If the whole sequence is labeled as inside or outside, the prediction is that it contains no membrane helices. It is probably not wise to interpret it as a prediction of location. The prediction gives the most probable location and orientation of transmembrane helices in the sequence. |
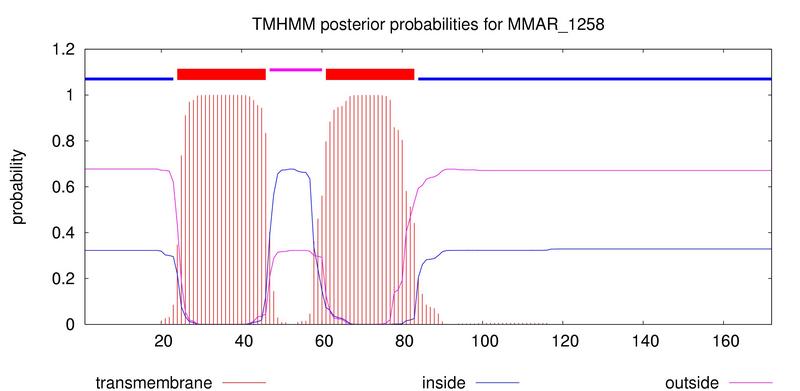
| Plot of probabilities |
| The plot shows the probabilities of inside/outside/TM helix. Possible weak TM helices are sometimes not predicted, and one can get an idea of the certainty of each segment in the prediction. |
| At the top of the plot (between 1 and 1.2) the best prediction is shown. |
| The plot is obtained by calculating the total probability that a residue sits in helix, inside, or outside summed over all possible paths through the model. Sometimes it seems like the plot and the prediction are contradictory, but that is because the plot shows probabilities for each residue, whereas the prediction is the over-all most probable structure. Therefore the plot should be seen as a complementary source of information. |
| Remarks |